- Introduction to the ERCC Data Coordination Center
- DCC Services
- Genboree Account
- What Can I Do with exRNA Profiling Data?
- The exRNA Atlas
- Submitting Your Data to the Atlas
- Information About Atlas Metadata
- Analyzing Your Own exRNA Data
- exRNA Tools
- DMRR/DCC Demos at Meetings
- Contact Us - Members of the DCC
Introduction to the ERCC Data Coordination Center¶
The Data Coordination Center (DCC) for the Extracellular RNA Communication Consortium (ERCC) is led by Prof. Aleksandar Milosavljevic
at the Bioinformatics Research Laboratory, Baylor College of Medicine, Houston, TX, USA.
- develop data and metadata standards for the ERCC
- establish data flow into the exRNA Atlas database
- develop tools for download, visualization and analysis of exRNA data
- integrate exRNA Atlas database with other relevant resources
DCC Services¶
Genboree Account¶
If you are a new user, please follow the steps below to obtain a Genboree account and access to all associated services.
- Sign up for a Genboree Account: You can sign up for a new Genboree account at http://www.genboree.org/. Click the Login/Register button in the top right corner and then select New Account from the dialog. Fill out the registration form with your details and hit Submit. You'll get an email asking you to confirm (typical signup/verification process).
- Log into the Genboree Commons and GenboreeKB: Next, you will need to sign in once to the Genboree Commons (used for exRNA related communications) and GenboreeKB (used for navigating exRNA metadata). You should use the username and password obtained from Step 1. Signing in once allows our system to recognize you so we can add you to the appropriate projects/sub-projects. Sign into the Genboree Commons at http://genboree.org/theCommons/login and the GenboreeKB at http://genboree.org/genboreeKB/login.
- Email the BRL exRNA Team: Finally, you will need to email BRL to gain access to the appropriate projects/sub-projects on the Genboree Commons and GenboreeKB. We will also provide a dedicated, shared directory for your lab on our FTP server so that your lab can upload submissions for the DMRR data and metadata processing pipeline. Please include your Genboree username and PI when you email us.
What Can I Do with exRNA Profiling Data?¶
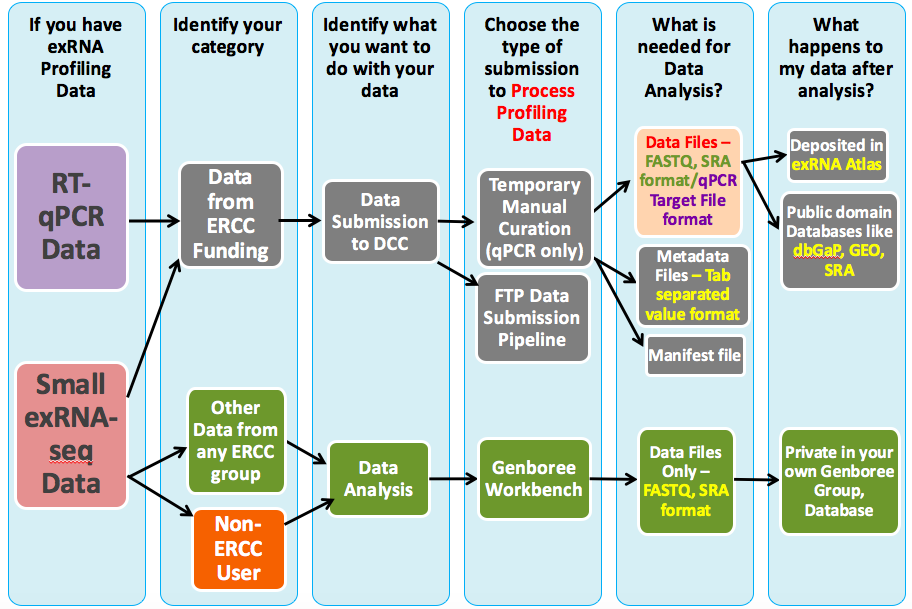
The exRNA Atlas¶
The exRNA Atlas is the data repository of the ERCC. It includes exRNA profiles derived from various biofluids and conditions and currently stores data profiled from small RNA sequencing assays and RT-qPCR assays.To learn more about the Atlas, you can read our tutorials:
- Using the ncRNA Search Bar
- Viewing Selected Biosamples in Grid via Faceted Charts
- Viewing Biosamples in Biosample Partition Grid
- Viewing Selected Biosamples in Grid via Linear Tree
- Downloading Data and Metadata from the exRNA Atlas
- Viewing exRNA Profiling Datasets
- Viewing Atlas Statistics
- Running Analyses and Viewing Analysis Results Using the exRNA Atlas
Submitting Your Data to the Atlas¶
You can also learn more about submitting your own data to the Atlas via our Data Submission to DCC using FTP Wiki page.
Information About Atlas Metadata¶
All Atlas metadata is stored in the Genboree KnowledgeBase, a MongoDB-backed database curation service.
Our metadata models follow the exRNA Metadata Standards developed by the Metadata and Data Standards (MADS) Working Group of the ERCC.
Analyzing Your Own exRNA Data¶
If you'd like to analyze your own data using the tools developed by the ERCC, you can use the Genboree Workbench to do so.
The Genboree Workbench is a web-based platform for performing data analysis. You can upload your data and perform various analyses using a "drag and drop" user interface.
To get started using the Genboree Workbench, you can view our collection of introductory materials.
exRNA Tools¶
Once you understand the basics of using the Workbench, you can start using the different ERCC tools to analyze your exRNA data:- The exceRpt Small RNA-seq Pipeline for exRNA Profiling can be used to analyze your small RNA-seq data (with long RNA-seq support coming soon)
- The Long RNA-seq Pipeline Using RSEQtools can be used to analyze your long RNA-seq data
- The KNIFE Circular and Linear Isoform Explorer can be used to detect circular and linear isoforms from RNA-seq data.
- The DESeq2 Differential Expression Analysis Tool can be used for differential expression analysis.
- The Target Interaction Finder Tool can be used for discovering miRNA-protein target interactions.
- The Pathway Finder Tool can be used to search for pathways either containing miRNAs of interest or protein targets of those miRNAs.
DMRR/DCC Demos at Meetings¶
- May 2014 - Demo of small and long RNA-Seq pipelines at the ERCC 2nd Investigators' Meeting, May 2014, at Bethesda, MD
- November 2014 - Demo of small RNA-seq pipeline and use cases presented at the ERCC 3rd Investigators' Meeting, November 2014, at Rockville, MD
- April 2015 - Demo of small RNA-seq pipeline and use cases presented at the ERCC 4th Investigators' Meeting and ISEV Annual Meeting, April 2015, at Bethesda, MD
- May 2015 - CIBR RNA-seq workshop - Demo of exceRpt small RNA processing pipeline, May 2015, at Baylor College of Medicine, Houston, TX
- November 2015 - Data Submission & Analysis Infrastructure at the DMRR - Talk at the ERCC 5th Investigators' Meeting, November 2015, at Rockville, MD
- April 2016 - DMRR Data Analysis and Bioinformatics Workshop - ERCC 6th Investigators' Meeting, April 2016, at Bethesda, MD
Contact Us - Members of the DCC¶
Prof. Aleksandar Milosavljevic - Principal Investigator
BRL Team - Point Person