Overview
- Using Pathway Finder to Perform a Search for Pathways Either Containing miRNAs of Interest or Protein Targets of Those miRNAs
- Introduction
- Instructional Video for Pathway Finder
- Preliminary Steps for Setting Up Any Analysis in the Genboree Workbench
- Create a Group for Your Analysis
- Create a Database for Your Analysis
- Upload your Data File(s)
- Step-by-step Instructions to Set Up Pathway Finder Submission
- Output Generated by Pathway Finder
- References and Attributions
Using Pathway Finder to Perform a Search for Pathways Either Containing miRNAs of Interest or Protein Targets of Those miRNAs¶
Introduction¶
The Pathway Finder tool takes a column of miRNA identifiers from an input text file and performs a search for pathways either containing miRNAs of interest or protein targets of those miRNAs.
A table of pathway results and an interactive pathway viewer are displayed in an output window after the tool successfully processes the input text file.
Instructional Video for Pathway Finder¶
Below, you can view an instructional video for using the Pathway Finder Tool on the Genboree Workbench:
Preliminary Steps for Setting Up Any Analysis in the Genboree Workbench¶
Create a Group for Your Analysis¶
What is a  Group? Group? |
A "Group" contains Databases and Projects and controls access to all content within. You control access to your Group(s), and who is a member of your group. You can also belong to multiple Groups (i.e. collaborators). |
This step is optional. You can also use your default/existing group.
Create a Database for Your Analysis¶
What is a  Database? Database? |
A "Database" contains Tracks, Lists, Sample Sets, Samples, and Files. Each database can be associated with a reference genome. |
This step is optional.
Because this tool doesn't create any on-disk output, you only need to create a Database if you want to upload your own input file(s) for processing.
If you are using an input file in a previously existing Database (like the "Pathway Finder - Example Data" Database in the "Examples and Test Data" Group),
then you do not need to create a new Database.
If you do create a Database, make sure that you pick the proper reference sequence genome (hg19, for example) when creating your database.
Note that we do not have an entry for hg38 or mm10 in our "Reference Sequence" list.
If your data is associated with either of these reference sequence genomes, follow the directions below.
Create a Database for hg38 or mm10
In order to create a database associated with hg38 or mm10, select the "User Will Upload" option for "Reference Sequence"
and provide appropriate values for the Species and Version text boxes as given below:
| Your Genome of Interest | Species | Version |
| Human genome hg38 | Homo sapiens | hg38 |
| Mouse genome mm10 | Mus musculus | mm10 |
Upload your Data File(s)¶
| What types of files can be uploaded? | The Pathway Finder tool accepts a single text file as input. This text file should have a column of miRNA identifiers as its first column. |
Step-by-step Instructions to Set Up Pathway Finder Submission¶
- Drag a single text file (with a first column consisting of miRNA identifiers) into the
Input Datapanel. - Select
Visualization»Pathway Finderfrom the Toolset menu. - Click Submit. When we finish processing your input file, an output window will pop up with pathway results and an interactive pathway viewer.
Output Generated by Pathway Finder¶
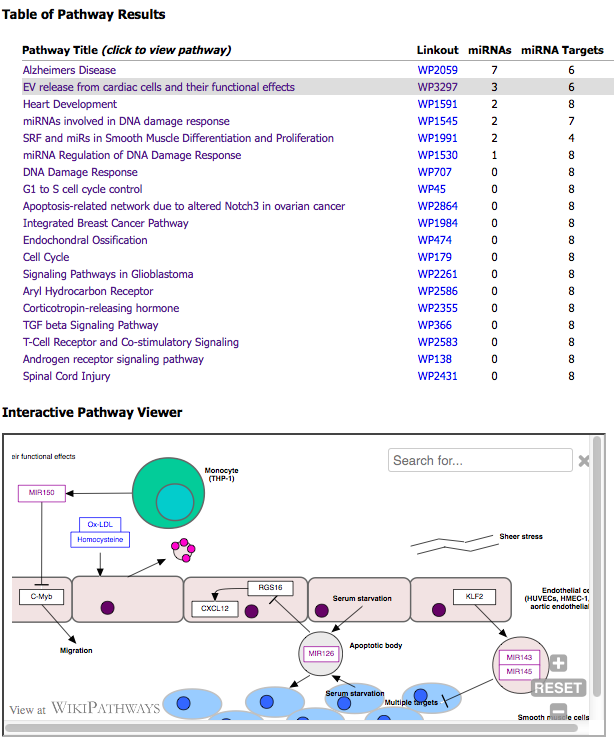
After your input file is successfully processed, you will be able to view a table of pathway results and an interactive pathway viewer.
The first column of the table lists a clickable pathway title that updates the viewer.
The second column lists pathway identifers that link to WikiPathways.org.
The list is sorted by the number of "miRNAs" (primary) and by "miRNA Targets" (secondary) found on each pathway.
The top 20 results are listed.
References and Attributions¶
- WikiPathways, source for curated pathways and miRNA content: Pico AR, et al. (2008) WikiPathways: Pathway Editing for the People. PLoS Biol 6(7). http://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060184
- miRTarBase source database for experimentally validated miRNA-protein target interactions: Chou et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. NAR, Database Issue, Vol 44(D1). http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702890/
- Pathway Finder tool was designed and implemented by Anders Riutta and Alexander Pico, and the video tutorial was produced by Kristina Hanspers, all at the Gladstone Institutes, San Francisco, CA.
- Integrated into the Genboree Workbench by William Thistlethwaite at the Bioinformatics Research Laboratory, Baylor College of Medicine, Houston, TX.
This tool has been deployed in the context of the exRNA Communication Consortium (ERCC).
Please contact William Thistlethwaite with questions or comments, or for help using it on your own data.
