Overview
- Generating miRNA-Protein Target Interaction Files for a Set of miRNA Identifiers Using Target Interaction Finder
- Introduction
- Instructional Video for Target Interaction Finder
- Preliminary Steps for Setting Up Any Analysis in the Genboree Workbench
- Create a Group for Your Analysis
- Create a Database for Your Analysis
- Upload your Data File(s)
- Step-by-step Instructions to Set Up Target Interaction Finder Submission
- Output Files Generated by Target Interaction Finder
- References and Attributions
Generating miRNA-Protein Target Interaction Files for a Set of miRNA Identifiers Using Target Interaction Finder¶
Introduction¶
This tool takes a column of miRNA identifiers from an input TXT file and generates miRNA-protein target interaction files in both a tabular CSV format and a network XGMML format.
The target interactions are sourced from miRTarBase 1, an experimentally validated miRNA-target interaction database.
The CSV and XGMML files can be imported into downstream tools, such as Cytoscape 2, for network analysis and visualization.
From a column of identifiers... to a network of interactions
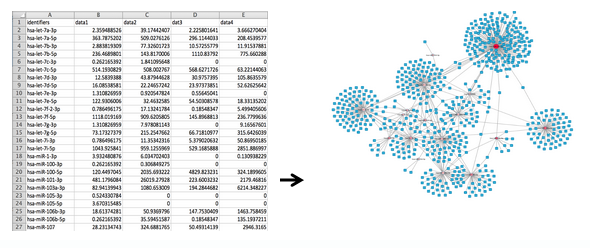
Instructional Video for Target Interaction Finder¶
Below, you can view an instruction video for using the Target Interaction Finder Tool on the Genboree Workbench:
Preliminary Steps for Setting Up Any Analysis in the Genboree Workbench¶
Create a Group for Your Analysis¶
What is a  Group? Group? |
A "Group" contains Databases and Projects and controls access to all content within. You control access to your Group(s), and who is a member of your group. You can also belong to multiple Groups (i.e. collaborators). |
This step is optional. You can also use your default/existing group.
Create a Database for Your Analysis¶
What is a  Database? Database? |
A "Database" contains Tracks, Lists, Sample Sets, Samples, and Files. Each database can be associated with a reference genome. |
This step is required.
Make sure that you pick the proper reference sequence genome (hg19, for example) when creating your database.
Note that we do not have an entry for hg38 or mm10 in our "Reference Sequence" list.
If your data is associated with either of these reference sequence genomes, follow the directions below.
Create a Database for hg38 or mm10
In order to create a database associated with hg38 or mm10, select the "User Will Upload" option for "Reference Sequence"
and provide appropriate values for the Species and Version text boxes as given below:
| Your Genome of Interest | Species | Version |
| Human genome hg38 | Homo sapiens | hg38 |
| Mouse genome mm10 | Mus musculus | mm10 |
Upload your Data File(s)¶
| What types of files can be uploaded? | The Target Interaction Finder tool accepts one or more text files as input. Each text file should have a column of miRNA identifiers as its first column. Your input text files can also be compressed, with each archive containing containing one or more input text files. For example, you could submit one archive containing 100 different input text files, or even three different archives containing different numbers of input text files. |
Step-by-step Instructions to Set Up Target Interaction Finder Submission¶
- Drag one or more text files (each with a first column consisting of miRNA identifiers) into the
Input Datapanel. These input file(s) can also be compressed, as mentioned above. - Drag a
Databaseto theOutput Targetspanel to store results. - Select
Visualization»Target Interaction Finderfrom the Toolset menu. - Fill in the analysis name for your tool job. We recommend keeping a timestamp in your analysis name!
- Submit your job. Upon completion of your job, you will receive an email.
- Download the results of your analysis from your
Database. The results data will end up under thetargetInteractionFinderfolder in theFilesarea of your output database.- Within that folder, your
Analysis Namewill be used as a sub-folder to hold the files generated by that run of the tool. - Open this sub-folder to see your results.
- Select any of the output files (explained in more detail below) and then click the link
Click to Download Filefrom theDetailspanel to download that output file.
- Within that folder, your
Output Files Generated by Target Interaction Finder¶
After your job successfully completes, you will be able to download 3 different output files:
- A .csv file that contains the following columns:
- queryid – the miRNA identifiers provided in the input file
- targeted – the protein identifier provided by miRTarBase, i.e., Entrez Gene
- pmid – pubmed identifier for article of evidence for interaction
- datasource – name of source database of miRNA-protein targets
- A .xgmml file that contains many annotations on both miRNA and protein nodes as well as on the interactions themselves.
- This file can be imported natively into network software tools such as Cytoscape.
- A summary.log file that contains summary information about the interactions found in the files above.
References and Attributions¶
- miRTarBase source database for experimentally validated miRNA-protein target interactions: Chou et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. NAR, Database Issue, Vol 44(D1). http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702890/
- Cytoscape tool for network visualization and analysis: Shannon, P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11). http://genome.cshlp.org/content/13/11/2498.full
- Target Interaction Finder tool written by Anders Riutta and Alexander Pico at the UCSF Gladstone Institute, San Francisco, CA.
- Integrated into the Genboree Workbench by William Thistlethwaite and Sai Lakshmi Subramanian at the Bioinformatics Research Laboratory, Baylor College of Medicine, Houston, TX.
This tool has been deployed in the context of the exRNA Communication Consortium (ERCC).
Please contact William Thistlethwaite with questions or comments, or for help using it on your own data.
