exRNA Toolset
The Genboree exRNA Toolset provides a web-based environment to conduct bioinformatics analyses on extracellular RNA profiling data.
The exRNA Toolset allows for analysis of both small and long RNA-seq data. Users can analyze their small RNA-seq data using the exceRpt Small RNA-seq Pipeline for exRNA Profiling, and they can analyze their long RNA-seq data using the RSEQtools pipeline. These tools can be found under the Transcriptome menu in the Genboree Workbench. Additionally, the exRNA Toolset includes tools for finding protein-target interaction networks and pathways for your miRNA targets. These tools can be found under the Visualization menu in the Workbench.
exRNA Data Analysis TutorialOverview of Tools
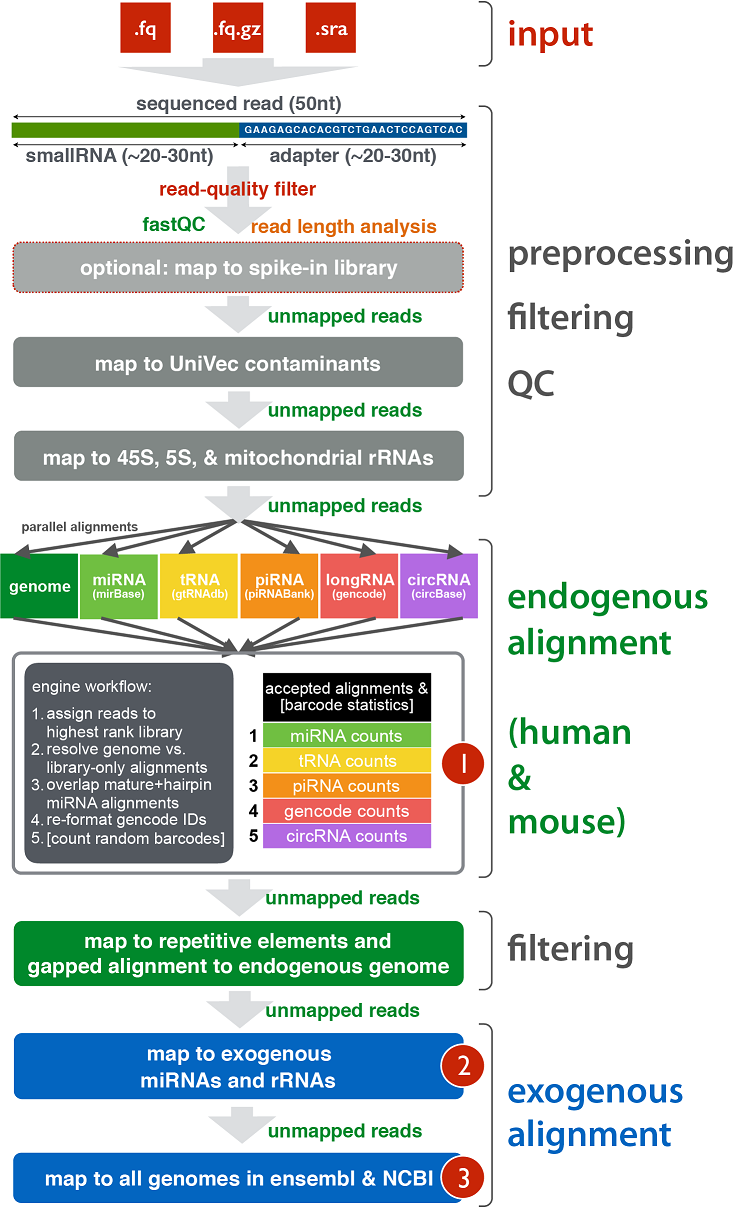
exceRpt Small RNA-seq Pipeline
The extracellular RNA processing tool (exceRpt) utilizes its own endogenous alignment and quantification engine to map and profile various small RNA libraries - these include miRNAs from miRBase, tRNAs from gtRNAdb, piRNAs from piRNABank, annotations from Gencode, and circRNAs from circBase. exceRpt also allows users to map to exogenous miRNAs and rRNAs as well as a large variety of exogenous genomes (bacteria, viruses, fungi, metazoa, etc.). Abundance estimates for each of the requested libraries are given as output, as are a variety of quality control metrics such as read-length distributions, summaries of reads mapped to each library, and detailed mapping information for each read mapped to each library. exceRpt also supports *N random barcode identification on the inner edges (3', 5', or both) of adapter sequences. These random barcodes help normalize the read-counts for amplification artifacts and serve as an alternative to the read-count for smallRNA quantitation.
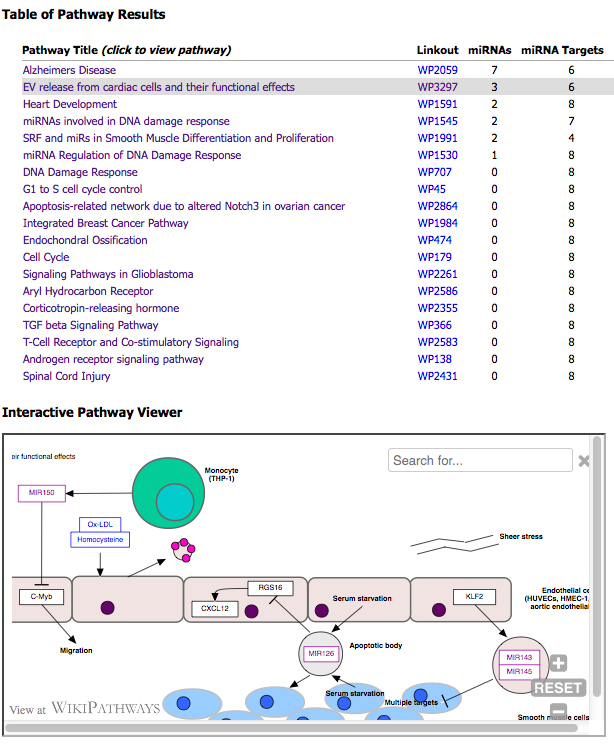
Pathway Finder
The Pathway Finder tool takes a column of miRNA identifiers from an input TXT file and performs a search for pathways either containing your miRNAs of interest or protein targets of your miRNAs. A table of pathway results and an interactive pathway viewer are displayed. The first column of the table lists a clickable pathway title that updates the viewer. The second column lists pathway identifiers that link to WikiPathways.org. The list is sorted by the number of "miRNAs" (primary) and by "miRNA Targets" (secondary) found on each pathway. The top 20 results are listed. Users can also access the Pathway Finder tool by using the ncRNA Search Bar on the exRNA Atlas.
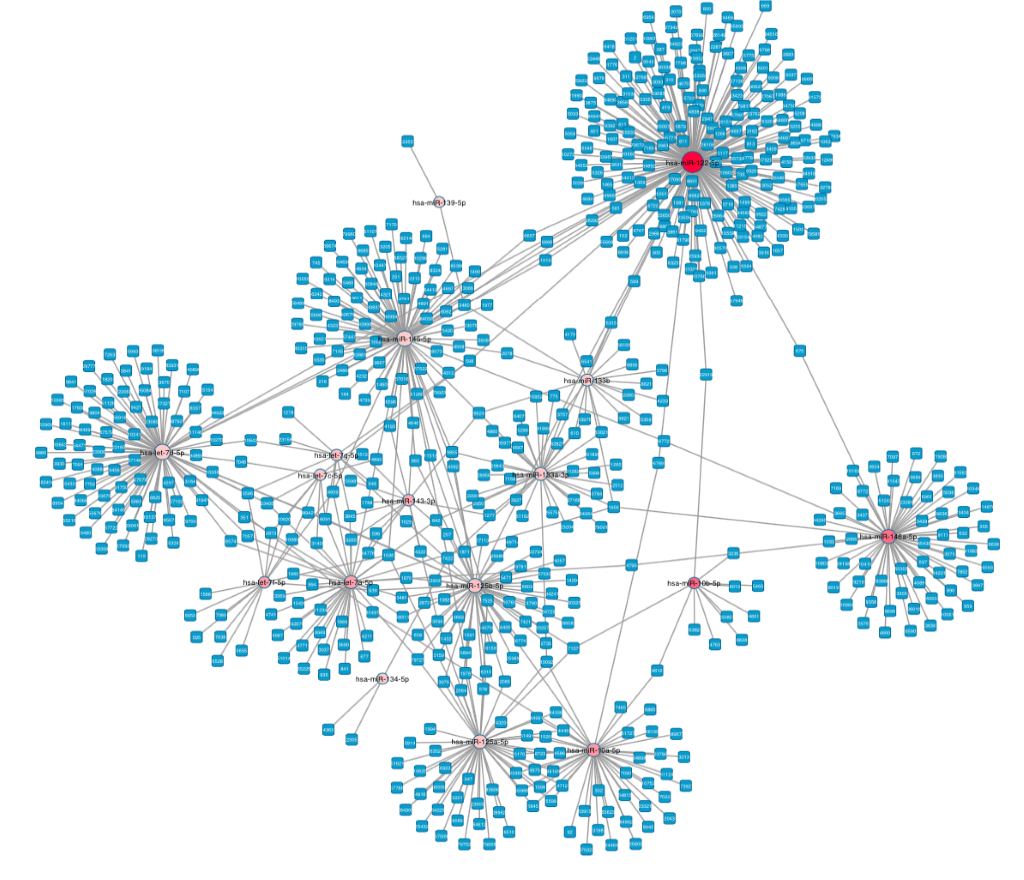
Target Interaction Finder
The Target Interaction Finder tool takes a column of miRNA identifiers from an input TXT file and generates miRNA-protein target interaction files in both a tabular CSV format and a network XGMML format. The target interactions are sourced from miRTarBase, an experimentally validated miRNA-target interaction database. The CSV and XGMML files can be imported into downstream tools, such as Cytoscape, for network analysis and visualization.
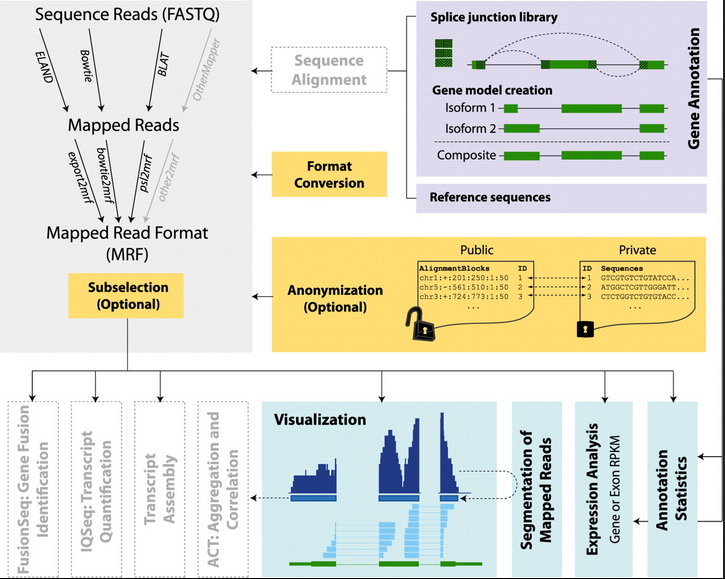
Long RNA-seq Pipeline
The long RNA-seq pipeline uses several well-established tools (FastQC, Bowtie, Samtools, and specific modules from RSEQtools) for long RNA-seq analysis. FastQC performs a quality check on the sequence reads, Bowtie maps the reads to the reference genome, and Samtools is used to post-process the aligned reads. Downstream analysis utilizes RSEQtools modules to perform gene-expression quantification, visualize signal tracks of mapped reads, calculate mapping bias, and compute annotation coverage.
Fold Change Calculation using DESeq2
The Fold Change Calculation using DESeq2 tool will compute fold change across samples for a given set of identifiers using DESeq2 (version 1.6.3). Currently, the tool allows you to test a given factor (disease, for example) across two different factor levels (control versus Alzheimer's disease, for example).
KNIFE (Known and Novel IsoForm Explorer) for Circular and Linear Isoforms
The KNIFE - Known and Novel IsoForm Explorer tool performs statistically based splicing detection for circular and linear isoforms from RNA-Seq data. The tool's statistical algorithm increases the sensitivity and specificity of circularRNA detection from RNA-Seq data by quantifying circular and linear RNA splicing events at both annotated and un-annotated exon boundaries.