-
Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. joural: Science. 2018 361: 6409. Doi: 10:1126/science.aar3146.
Onuchic V, Lurie E, Carrero I, Pawliczek P, Patel R, Rozowsky J, Galeev T, Huang Z, Altshuler R, Zhang Z, Harris R, Coarfa C, Ashmore L, Bertol J, Fakhouri W, Yu F, Kellis M, Gerstein M, and Milosavljevic A.
-
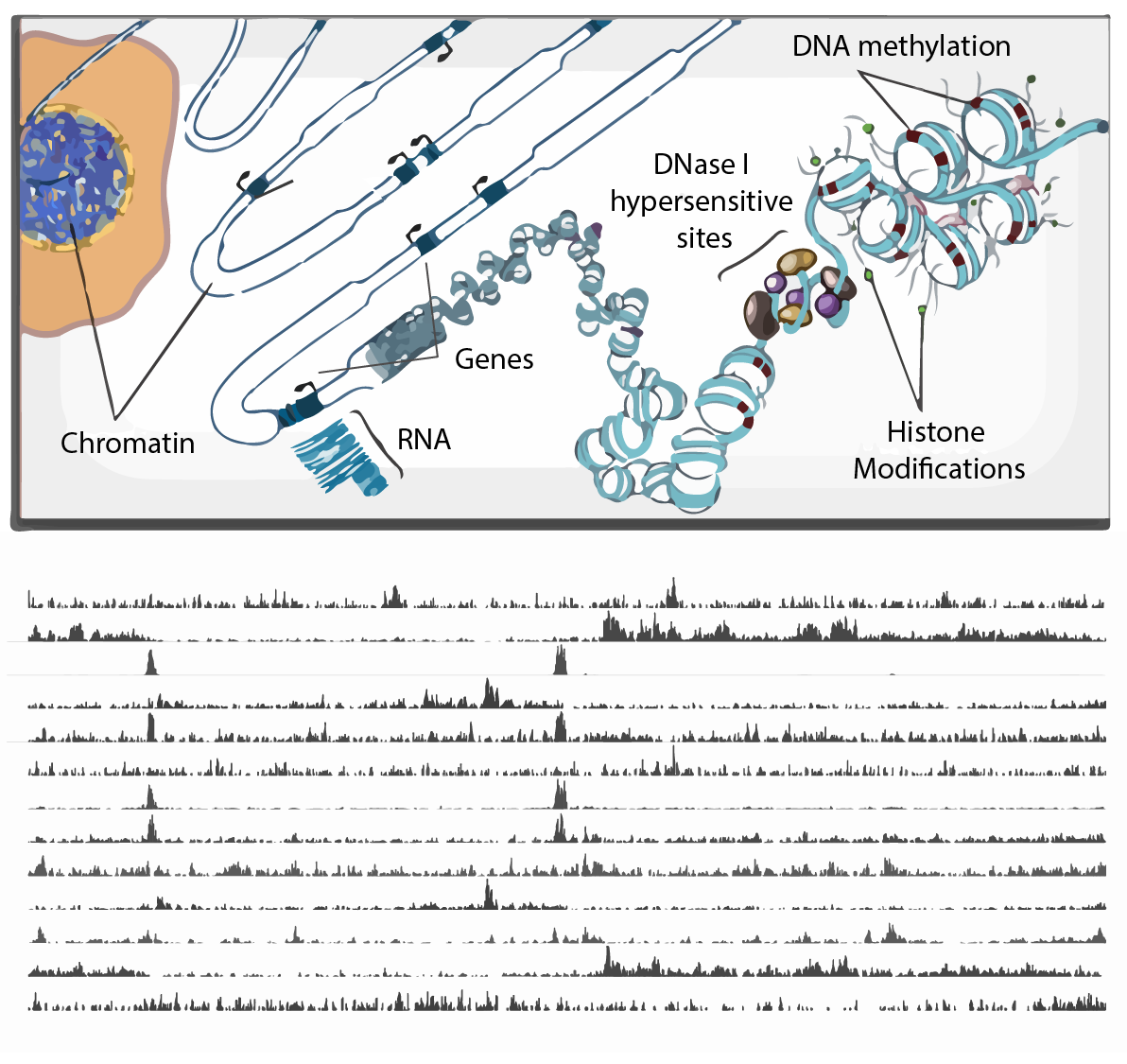
The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery
International Human Epigenome Consortium
Cell. doi: 10.1016/j.cell.2016.11.007 (2016)
PMID: 27863232
-
Epigenomic Deconvolution of Breast Tumors Reveals Metabolic Coupling between Constituent Cell Types
Onuchic V, Hartmaier RJ, Boone DN, Samuels ML, Patel RY, White WM, Garovic VD, Oesterreich S, Roth ME, Lee AV, Milosavljevic A.
Cell Rep. doi: 10.1016/j.celrep.2016.10.057 (2016)
PMID: 27851969
-
Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs
Amin V, Harris RA, Onuchic V, Jackson A, Charnecki T, Paithankar S, Subramanian S, Riehle K, Coarfa C, Milosavljevic A.
Nat. Commun. 6:6370 doi: 10.1038/ncomms7370 (2015)
PMID: 25691256
-
Integrative analysis of 111 reference human epigenomes
Roadmap Epigenomics Consortium et. al.
Nature doi: 10.1038/nature14248 (2015)
PMID: 25693563
-
Analysis of interactions between the epigenome and structural mutability of the genome using Genboree Workbench tools
Coarfa C, Pichot C, Jackson A, Tandon A, Amin V, Raghuraman S, Paithankar S, Lee AV, McGuire SE, Milosavljevic A.
BMC Bioinformatics. 2014;15 Suppl 7:S2. doi: 10.1186/1471-2105-15-S7-S2. Epub 2014 May 28.
PMID: 25080362
-
Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing
Kunde-Ramamoorthy G, Coarfa C, Laritsky E, Kessler NJ, Harris RA, Xu M, Chen R, Shen L, Milosavljevic A, Waterland RA.
Nucleic Acids Res. 2014 Apr;42(6):e43. doi: 10.1093/nar/gkt1325. Epub 2014 Jan 3.
PMID: 24391148
-
Recommendations for the design and analysis of epigenome-wide association studies
Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Houseman EA, Izzi B, Kelsey KT, Meissner A, Milosavljevic A, Siegmund KD, Bock C, Irizarry RA.
Nat Methods. 2013 Oct;10(10):949-55. doi: 10.1038/nmeth.2632. Review.
PMID: 24076989
-
Confounding by repetitive elements and CpG islands does not explain the association between hypomethylation and genomic instability
Harris RA, Shaw C, Li J, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Chinault AC, Gambin T, Gambin A, Lupski JR, Milosavljevic A.
PLoS Genet. 2013;9(2):e1003333. doi: 10.1371/journal.pgen.1003333. Epub 2013 Feb 28. No abstract available.
PMID: 23468659
-
Spark: a navigational paradigm for genomic data exploration
Nielsen CB, Younesy H, O'Geen H, Xu X, Jackson AR, Milosavljevic A, Wang T, Costello JF, Hirst M, Farnham PJ, Jones SJ.
Genome Res. 2012 Nov;22(11):2262-9. doi: 10.1101/gr.140665.112. Epub 2012 Sep 7.
PMID: 22960372
-
Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood
Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Saraj S, Dobson R, Schalkwyk LC, Mill J.
Genome Biol. 2012 Jun 15;13(6):R43. doi: 10.1186/gb-2012-13-6-r43.
PMID: 22703893
-
Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome
Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, Chinault AC, Gambin T, Gambin A, Lupski JR, Milosavljevic A.
PLoS Genet. 2012;8(5):e1002692. doi: 10.1371/journal.pgen.1002692. Epub 2012 May 17.
PMID: 22615578
-
Emerging patterns of epigenomic variation
Milosavljevic A
Trends in Genetics, 2011 Jun;27(6):242-50
PMID: 21507501
-
Putting epigenome comparison into practice
Milosavljevic A
Nat Biotechnol., 2010 Oct;28(10):1053-6
PMID: 20944597
-
The NIH Roadmap Epigenomics Mapping Consortium
Bernstein BE, Stamatoyannopoulus JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA
Nat Biotechnol., 2010 Oct;28(10):1045-8
PMID: 20944595
-
Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications
Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O'Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF
Nat Biotechnol., 2010 Oct;28(10):1097-105
PMID: 20852635