- Viewing exRNA Profiling Datasets
- Dataset Submissions Table
- Datasets Page
- RNA Profile Grid
- Sample Metadata Grid
Viewing exRNA Profiling Datasets¶
All profiles that are submitted to the exRNA Atlas are part of a dataset.
Each dataset is associated with a given study that focuses on some topic (detection of biomarkers associated with gastric cancer, for example).
There are two different ways of viewing datasets on the exRNA Atlas.
Dataset Submissions Table¶
First, on the Atlas home page, you can find the Dataset Submissions table.
This table provides a summary-level description for each dataset submission to the Atlas.
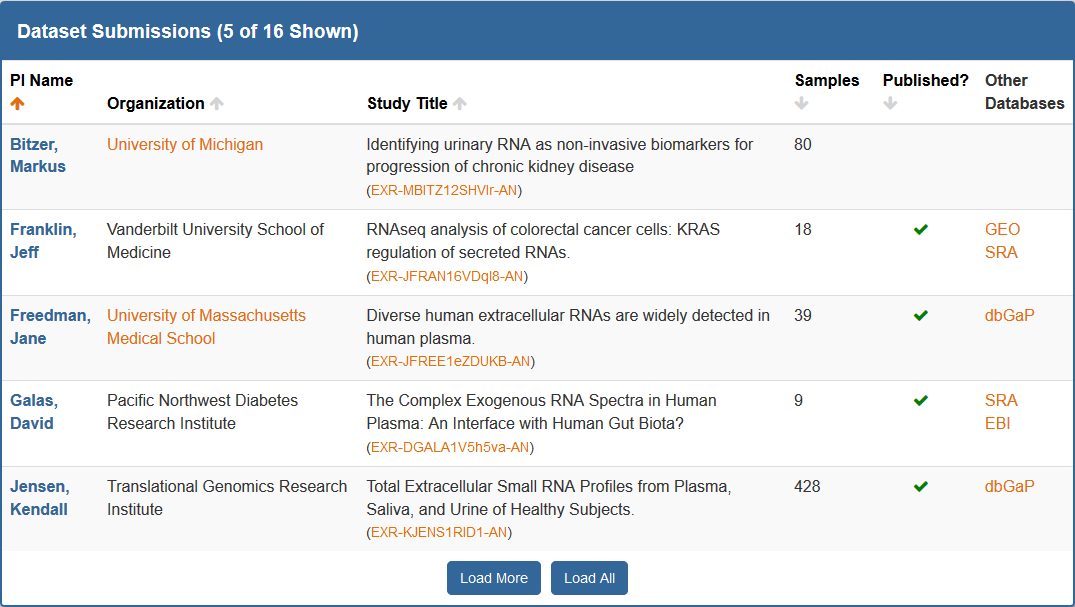
The table, by default, is organized by PI (last) name, but you can sort (ascending or descending) by most of the columns.
Clicking the analysis ID for a given dataset in the Study Title column will take you to its card on the stand-alone Datasets page (described below).
Clicking the green check mark for a given dataset in the Published? column will open the publication associated with that dataset.
Clicking the name of an external database (dbGaP, GEO, SRA) for a given dataset in the Other Databases column will open the associated page for that dataset in the external database.
You can click Load More to load an additional 5 datasets, or click Load All to load all datasets at once.
If you want the table to return to default, you can then click the Return to Default button (only available once you've loaded additional datasets).
Datasets Page¶
If you want to view datasets in more detail, you can visit the stand-alone Datasets page.You can reach this page in three different ways:
- Click the Datasets button in the navigation bar at the top of any Atlas page
- Click the exRNA Profiling Datasets link in the Browse exRNA Profiles - Alternative Options panel near the bottom of the Atlas home page
- Click the analysis ID associated with a given dataset in the Dataset Submissions table
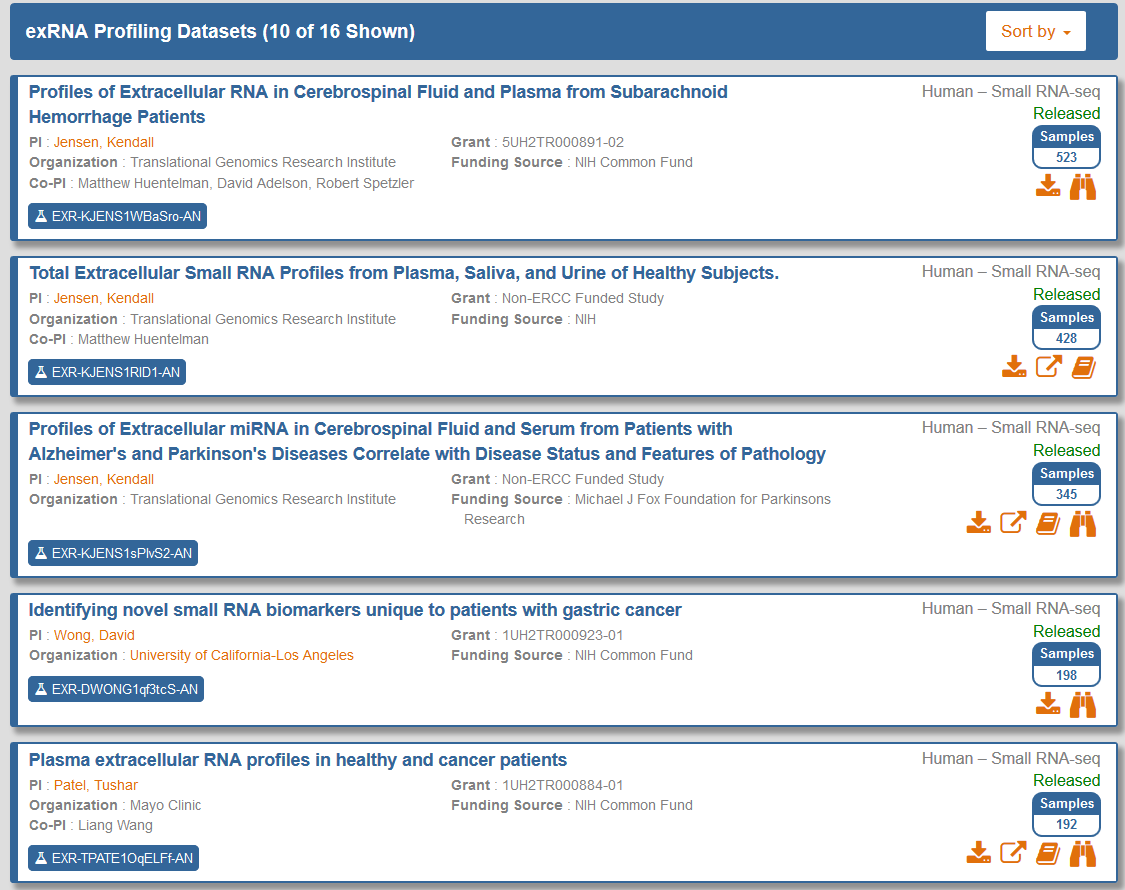
- The Analysis ID in the lower left corner will open an RNA profile grid for that dataset.
- For RNA-seq profiles, this grid will contain different read counts from various stages of mapping in the exceRpt pipeline.
- For qPCR profiles, this grid will contain sample metadata.
- The Samples badge on the right side will open a grid containing sample metadata for that dataset.
- The
 button will bring up a pop-over window that contains various downloads associated with the dataset.
button will bring up a pop-over window that contains various downloads associated with the dataset.
- The
 button will download a PDF containing different diagnostic plots for the dataset.
button will download a PDF containing different diagnostic plots for the dataset. - The
 button will download a table of the different raw (not normalized) miRNA read counts for the dataset.
button will download a table of the different raw (not normalized) miRNA read counts for the dataset. - The
 button will download a text file containing the exogenous genomic taxonomy's cumulative read counts for the dataset.
button will download a text file containing the exogenous genomic taxonomy's cumulative read counts for the dataset. - The
 button will download a text file containing the exogenous ribosomal RNA taxonomy's cumulative read counts for the dataset.
button will download a text file containing the exogenous ribosomal RNA taxonomy's cumulative read counts for the dataset. - The
 button will download an archive containing a large assortment of different summary files for this dataset.
button will download an archive containing a large assortment of different summary files for this dataset.
- The
- The
 button will bring up a pop-over window that contains links to external references to the dataset.
button will bring up a pop-over window that contains links to external references to the dataset.
- Examples include dbGaP, GEO, BioProject, and ArrayExpress.
- The
 button will bring up a pop-over window that contains links to PubMed articles associated with the dataset.
button will bring up a pop-over window that contains links to PubMed articles associated with the dataset. - The
 button will open up an overview page for the dataset on BioGPS, a gene annotation portal that will allow you to visualize counts for different miRNA species present in the dataset.
button will open up an overview page for the dataset on BioGPS, a gene annotation portal that will allow you to visualize counts for different miRNA species present in the dataset.
Note that not all options will be available for each card.
RNA Profile Grid¶
By clicking the Analysis ID associated with a given dataset, you can pull up a grid that contains read counts for that dataset.
The grid will also contain various downloads for each sample in the dataset.
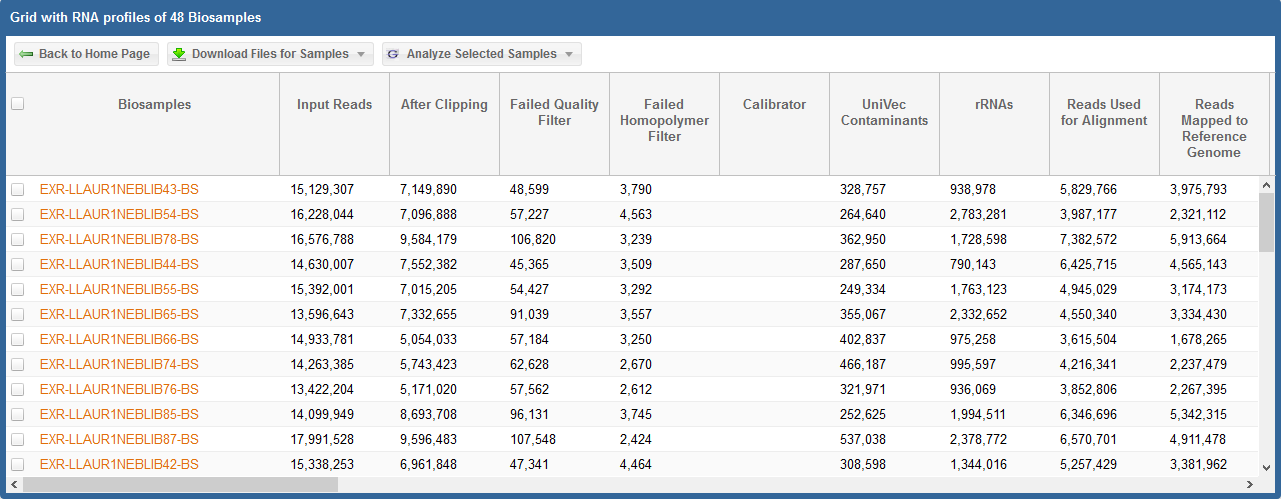
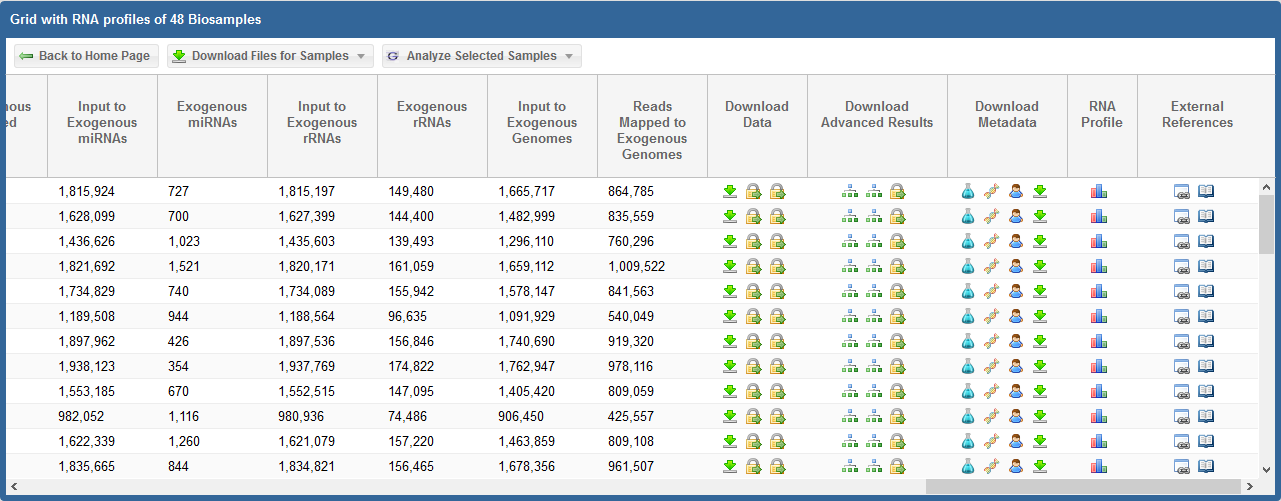
In the first picture above, we see the read counts associated with different exceRpt mapping stages for each sample.
In the second picture above, we see the following information and links:
Download Data
- For all profiles, click the
 icon to download the "core processed results" associated with the sample.
icon to download the "core processed results" associated with the sample.
- For RNA-seq profiles, this download will be the exceRpt processed core results archive.
This archive will contain mapped read counts from all three stages of exceRpt (endogenous, exogenous miRNA and rRNA, and exogenous genomes). - For qPCR profiles, this download will be the qPCR Targets file. The file will contain different miRNA targets and associated Ct values for those targets.
- For RNA-seq profiles, this download will be the exceRpt processed core results archive.
- For RNA-seq profiles, click the
 icon to download the full results (alignments) for the first two stages of exceRpt (endogenous alignment and exogenous miRNA and rRNA alignment).
icon to download the full results (alignments) for the first two stages of exceRpt (endogenous alignment and exogenous miRNA and rRNA alignment). - For RNA-seq profiles, click the
 icon to download the original FASTQ source file.
icon to download the original FASTQ source file. - If you see
 , this icon means that the data is restricted access and is currently under the protected period (embargo).
, this icon means that the data is restricted access and is currently under the protected period (embargo).
The embargo on this dataset will end 12 months after the time the data was submitted to the DCC. View the ERC Consortium Data Access Policy for more details. - If you see
 , this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
, this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
You can click the icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
You can then request access through those external databases.
Download Advanced Results
- For RNA-seq profiles, click the
 icon to download the taxonomy tree (either exogenous ribosomal RNA or exogenous genomic reads) created by exceRpt.
icon to download the taxonomy tree (either exogenous ribosomal RNA or exogenous genomic reads) created by exceRpt. - For RNA-seq profiles, click the
 icon to download the full results (alignments) for the third stage of exceRpt (exogenous genomic alignment).
icon to download the full results (alignments) for the third stage of exceRpt (exogenous genomic alignment). - If you see
 , this icon means that the data is restricted access and is currently under the protected period (embargo).
, this icon means that the data is restricted access and is currently under the protected period (embargo).
The embargo on this dataset will end 12 months after the time the data was submitted to the DCC. View the ERC Consortium Data Access Policy for more details. - If you see
 , this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
, this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
You can click the icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
You can then request access through those external databases.
Download Metadata
- Click the
 icon to download the biosample metadata document associated with the biosample.
icon to download the biosample metadata document associated with the biosample.
You can also view the document in GenboreeKB (our UI for viewing metadata) by clicking the biosample's accession ID in the Biosample Metadata Accession column. - Click the
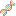 icon to download the experiment metadata document associated with the biosample.
icon to download the experiment metadata document associated with the biosample. - Click the
 icon to download the donor metadata document associated with the biosample.
icon to download the donor metadata document associated with the biosample. - Click the
 icon to download a file containing all three metadata documents (biosample, donor, and experiment) associated with the biosample.
icon to download a file containing all three metadata documents (biosample, donor, and experiment) associated with the biosample.
RNA Profile
- Click the
 icon to view a histogram of read counts mapped to various libraries.
icon to view a histogram of read counts mapped to various libraries.
External References
- Click the
 icon to view information about external databases associated with the biosample.
icon to view information about external databases associated with the biosample.
If the biosample can be found in any external databases (SRA, dbGaP, GEO, etc.), then a link is provided.
If the biosample is still embargoed, then information about the embargo period is displayed, along with a link to the ERCC data access policy. - Click the
 icon to open the PubMed page associated with the biosample.
icon to open the PubMed page associated with the biosample.
If there is no PubMed page, you will get a pop-up alerting you that no references could be found.
There are three additional buttons at the top of the grid.
First, the Back to Home Page button will take you back to the exRNA Atlas landing page.
Second, the Download All Samples button will allow you to download result files in bulk.
To learn more about this option, view this tutorial.
Third, the Analyze Selected Samples button will allow you to take samples from the exRNA Atlas and feed them into downstream and comparative analysis tools.
To learn more about this option, view this tutorial.
Sample Metadata Grid¶
By clicking the Samples badge associated with a given dataset, you can pull up a grid that contains sample metadata for that dataset.
In the first picture above (which displays the first half of the grid), we see each biosample's name as well as some key metadata properties
of each biosample (Condition, Anatomical Location, Biofluid Name, and exRNA Source).
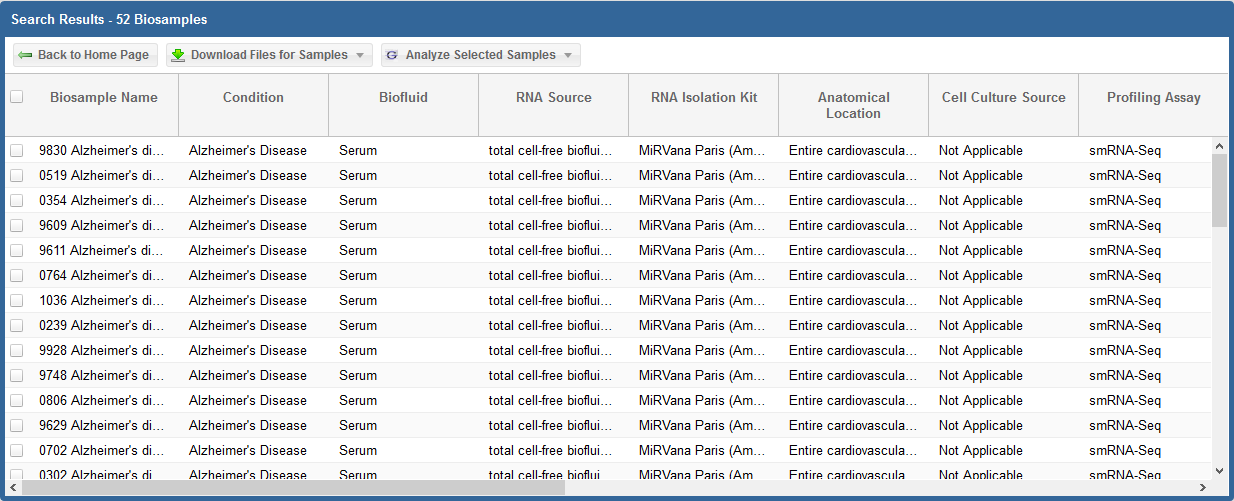
In the second picture above (which displays the second half of the grid), we see the following information and links:
ERCC Quality Standards?
- The grid will display ERCC quality standard metrics for each sample.
The "Meets Standards?" column will clearly indicate whether the sample meets the required quality thresholds: "YES", "NO", or "NA".
A value of "NA" indicates that we are currently reevaluating that sample's quality.
You can view the ERC Consortium QC Standards page to learn more about the QC standards used.
Download Data
- For all profiles, click the
 icon to download the "core processed results" associated with the sample.
icon to download the "core processed results" associated with the sample.
- For RNA-seq profiles, this download will be the exceRpt processed core results archive.
This archive will contain mapped read counts from all three stages of exceRpt (endogenous, exogenous miRNA and rRNA, and exogenous genomes). - For qPCR profiles, this download will be the qPCR Targets file. The file will contain different miRNA targets and associated Ct values for those targets.
- For RNA-seq profiles, this download will be the exceRpt processed core results archive.
- For RNA-seq profiles, click the
 icon to download the full results (alignments) for the first two stages of exceRpt (endogenous alignment and exogenous miRNA and rRNA alignment).
icon to download the full results (alignments) for the first two stages of exceRpt (endogenous alignment and exogenous miRNA and rRNA alignment). - For RNA-seq profiles, click the
 icon to download the original FASTQ source file.
icon to download the original FASTQ source file. - If you see
 , this icon means that the data is restricted access and is currently under the protected period (embargo).
, this icon means that the data is restricted access and is currently under the protected period (embargo).
The embargo on this dataset will end 12 months after the time the data was submitted to the DCC. View the ERC Consortium Data Access Policy for more details. - If you see
 , this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
, this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
You can click the icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
You can then request access through those external databases.
Download Advanced Results
- For RNA-seq profiles, click the
 icon to download the taxonomy tree (either exogenous ribosomal RNA or exogenous genomic reads) created by exceRpt.
icon to download the taxonomy tree (either exogenous ribosomal RNA or exogenous genomic reads) created by exceRpt. - For RNA-seq profiles, click the
 icon to download the full results (alignments) for the third stage of exceRpt (exogenous genomic alignment).
icon to download the full results (alignments) for the third stage of exceRpt (exogenous genomic alignment). - If you see
 , this icon means that the data is restricted access and is currently under the protected period (embargo).
, this icon means that the data is restricted access and is currently under the protected period (embargo).
The embargo on this dataset will end 12 months after the time the data was submitted to the DCC. View the ERC Consortium Data Access Policy for more details. - If you see
 , this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
, this icon means that the data is deposited (or will be soon) into a controlled access archive like dbGaP.
You can click the icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
icon under the Actions column to view any available links to controlled access archive(s) that contain data for the relevant biosample.
You can then request access through those external databases.
Download Metadata
- Click the
 icon to download the biosample metadata document associated with the biosample.
icon to download the biosample metadata document associated with the biosample.
You can also view the document in GenboreeKB (our UI for viewing metadata) by clicking the biosample's accession ID in the Biosample Metadata Accession column. - Click the
 icon to download the experiment metadata document associated with the biosample.
icon to download the experiment metadata document associated with the biosample. - Click the
 icon to download the donor metadata document associated with the biosample.
icon to download the donor metadata document associated with the biosample. - Click the
 icon to download a file containing all three metadata documents (biosample, donor, and experiment) associated with the biosample.
icon to download a file containing all three metadata documents (biosample, donor, and experiment) associated with the biosample.
RNA Profile
- Click the
 icon to view a histogram of read counts mapped to various libraries.
icon to view a histogram of read counts mapped to various libraries.
External References
- Click the
 icon to view information about external databases associated with the biosample.
icon to view information about external databases associated with the biosample.
If the biosample can be found in any external databases (SRA, dbGaP, GEO, etc.), then a link is provided.
If the biosample is still embargoed, then information about the embargo period is displayed, along with a link to the ERCC data access policy. - Click the
 icon to open the PubMed page associated with the biosample.
icon to open the PubMed page associated with the biosample.
If there is no PubMed page, you will get a pop-up alerting you that no references could be found.
There are three additional buttons at the top of the grid.
First, the Back to Home Page button will take you back to the exRNA Atlas landing page.
Second, the Download All Samples button will allow you to download result files in bulk.
To learn more about this option, view this tutorial.
Third, the Analyze Selected Samples button will allow you to take samples from the exRNA Atlas and feed them into downstream and comparative analysis tools.
To learn more about this option, view this tutorial.